单细胞转录组实战06: pySCENIC转录因子分析(可视化)
原创
from pathlib import Path
import operator
import cytoolz
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import scanpy as sc
from pyscenic.utils import load_motifs
from pyscenic.aucell import aucell
from pyscenic.binarization import binarize
from pyscenic.plotting import plot_binarization, plot_rss
from pyscenic.transform import df2regulons
import bioquest as bq #https://jihulab.com/BioQuest/bioquestOUTPUT_DIR='output/05.SCENIC'
Path(OUTPUT_DIR).mkdir(parents=True,exist_ok=True)adata = sc.read_h5ad('output/03.inferCNV/adata.h5')
adata_raw = adata.raw.to_adata()自定义函数
def display_logos(df: pd.DataFrame,
top_target_genes: int = 3,
base_url: str = "http://motifcollections.aertslab.org/v10/logos/"
,column_name_logo = "MotifLogo"
,column_name_id = "MotifID"
, column_name_target = "TargetGenes"
:param df:
:param base_url:
# Make sure the original dataframe is not altered.
df = df.copy()
# Add column with URLs to sequence logo.
def create_url(motif_id):
return '<img src="{}{}.png" style="max-height:124px;"></img>'.format(base_url, motif_id)
df[("Enrichment", column_name_logo)] = list(map(create_url, df.index.get_level_values(column_name_id)))
# Truncate TargetGenes.
def truncate(col_val):
return sorted(col_val, key=op.itemgetter(1))[:top_target_genes]
df[("Enrichment", column_name_target)] = list(map(truncate, df[("Enrichment", column_name_target)]))
MAX_COL_WIDTH = pd.get_option('display.max_colwidth')
pd.set_option('display.max_colwidth', -1)
display(HTML(df.head().to_html(escape=False)))
pd.set_option('display.max_colwidth', MAX_COL_WIDTH)def fetch_logo(regulon, base_url = "http://motifcollections.aertslab.org/v10/logos/"):
for elem in regulon.context:
if elem.endswith('.png'):
return '<img src="{}{}" style="max-height:124px;"></img>'.format(base_url, elem)
return ""binarization Visualisation
auc_mtx=pd.read_csv(OUTPUT_DIR+"/aucell.csv", index_col=0)
bin_mtx = pd.read_csv(OUTPUT_DIR+"/bin_mtx.csv", index_col=0)
thresholds = pd.read_csv(OUTPUT_DIR+"/thresholds.csv", index_col=0).threshold
# 删除基因后的(+)
auc_mtx.columns = bq.st.removes(string=auc_mtx.columns, pattern=r'\(\+\)')
bin_mtx.columns = bq.st.removes(string=bin_mtx.columns, pattern=r'\(\+\)')
thresholds.index = bq.st.removes(string=thresholds.index, pattern=r'\(\+\)')- AUC
regulon双峰图,以及红线表示二值化的阈值
auc_sum = auc_mtx.apply(sum,axis=0).sort_values(ascending=False)
fig, axes = plt.subplots(1, 5, figsize=(8, 2), dpi=100)
for x,y in enumerate(axes):
plot_binarization(auc_mtx, auc_sum.index[x], thresholds[auc_sum.index[x]], ax=y)
plt.tight_layout()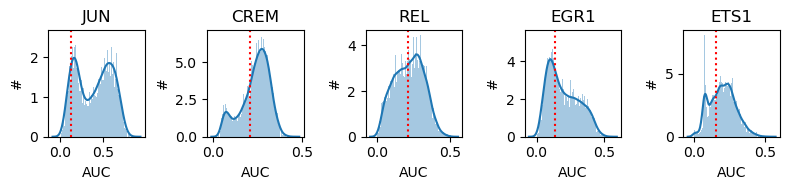
- 二值化热图
cell_type_key = "CellTypeS2"cell_type_color_lut = dict(zip(adata.obs[cell_type_key].dtype.categories, adata.uns[f'{cell_type_key}_colors']))
cell_id2cell_type_lut = adata.obs[cell_type_key].to_dict()
bw_palette = sns.xkcd_palette(["white", "black"])sns.set()
sns.set(font_scale=1.0)
sns.set_style("ticks", {"xtick.minor.size": 1, "ytick.minor.size": 0.1})
g = sns.clustermap(
data=bin_mtx.T,
col_colors=auc_mtx.index.map(cell_id2cell_type_lut).map(cell_type_color_lut),
cmap=bw_palette, figsize=(20,20)
g.ax_heatmap.set_xticklabels([])
g.ax_heatmap.set_xticks([])
g.ax_heatmap.set_xlabel('Cells')
g.ax_heatmap.set_ylabel('Regulons')
g.ax_col_colors.set_yticks([0.5])
g.ax_col_colors.set_yticklabels(['Cell Type'])
g.cax.set_visible(False)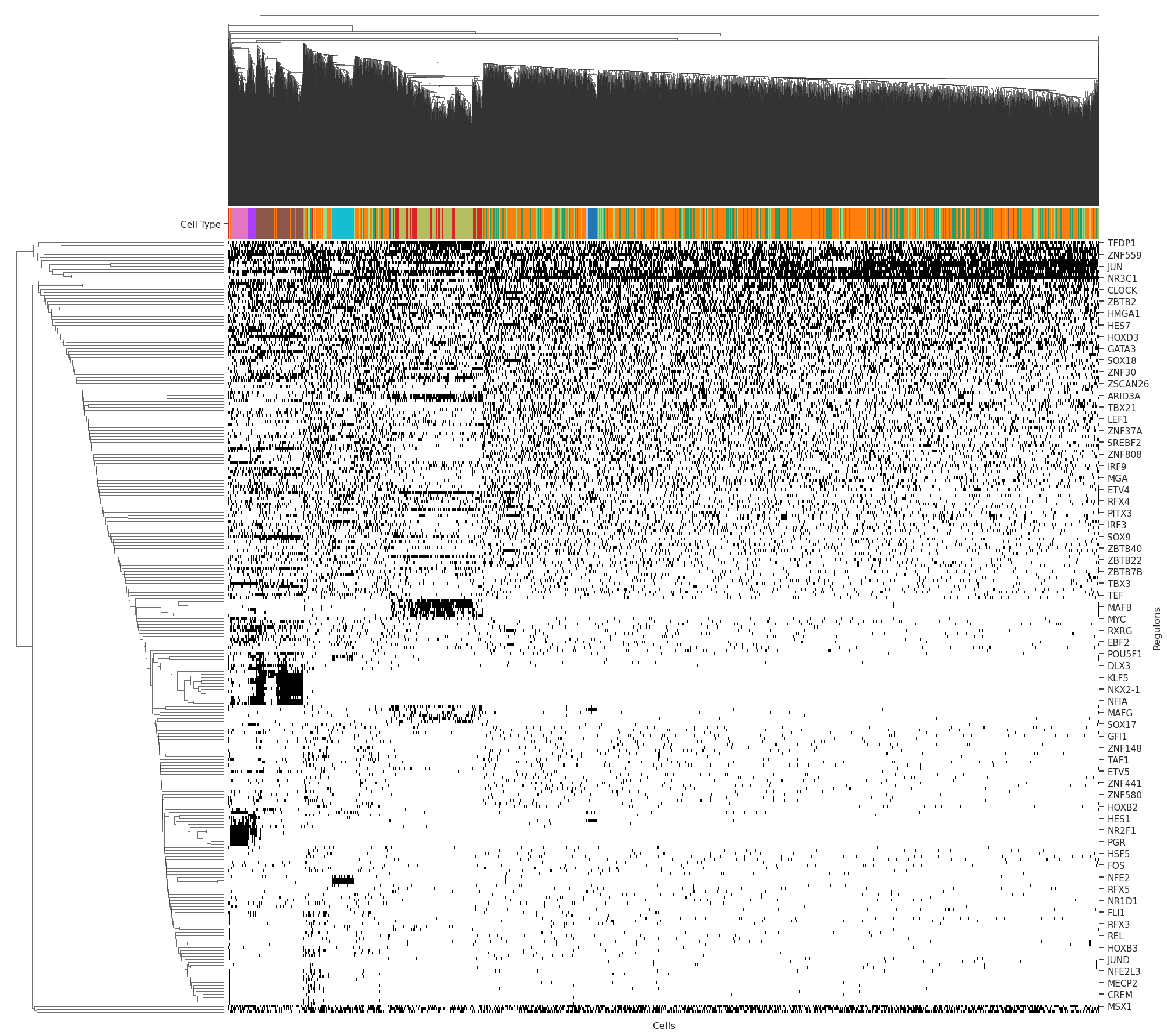
DNA序列logo图
df_regulons = pd.DataFrame(data=[list(map(operator.attrgetter('name'), regulons)),
list(map(len, regulons)),
list(map(fetch_logo, regulons))],
index=['name', 'count', 'logo']).TMAX_COL_WIDTH = pd.get_option('display.max_colwidth')
pd.set_option('display.max_colwidth', -1)
import IPython.display
IPython.display.display(IPython.display.HTML(df_regulons.iloc[[2,5,7],:].to_html(escape=False)))
pd.set_option('display.max_colwidth', MAX_COL_WIDTH)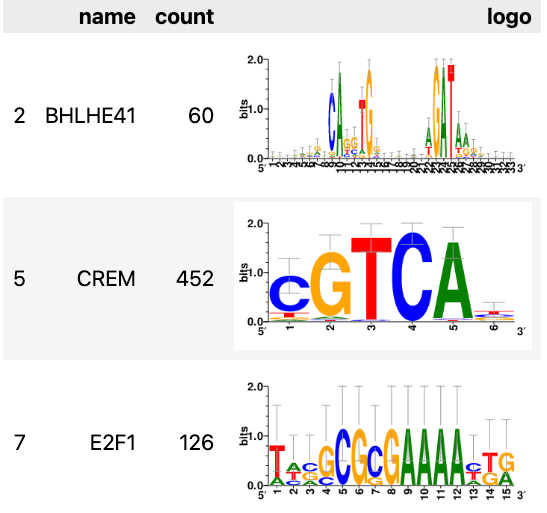
UMAP
基于"X_aucell"聚类
from pyscenic.export import add_scenic_metadata
add_scenic_metadata(adata, auc_mtx, regulons);sc.tl.umap(adata,init_pos="X_aucell")
sc.pl.umap(adata,color=cell_type_key)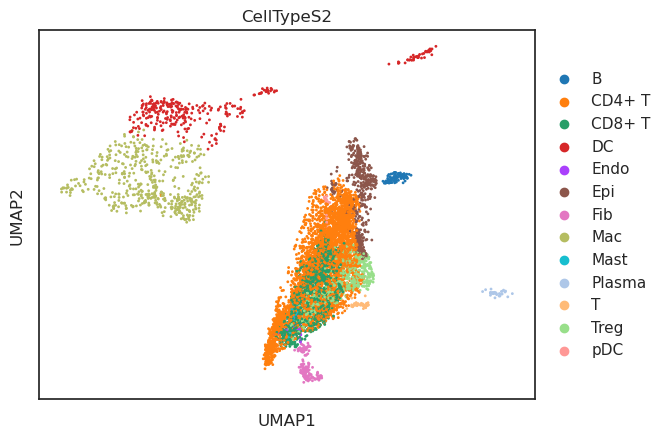
细胞特异 REGULATORS
df_scores=bq.tl.select(adata.obs,columns=[cell_type_key],pattern=r'^Regulon\(')
df_results = ((df_scores.groupby(by=cell_type_key).mean() - df_scores[df_scores.columns[1:]].mean())/ df_scores[df_scores.columns[1:]].std()).stack().reset_index().rename(columns={'level_1': 'regulon', 0:'Z'})
df_results['regulon'] = list(map(lambda s: s[8:-1], df_results.regulon))
df_results[(df_results.Z >= 3.0)].sort_values('Z', ascending=False).head()df_heatmap = pd.pivot_table(data=df_results[df_results.Z >= 3.0].sort_values('Z', ascending=False),index=cell_type_key, columns='regulon', values='Z')
fig, ax1 = plt.subplots(1, 1, figsize=(10, 8))
sns.heatmap(df_heatmap, ax=ax1, annot=True, fmt=".1f", linewidths=.7, cbar=False, square=True, linecolor='gray',cmap="viridis", annot_kws={"size": 8})
ax1.set_ylabel('');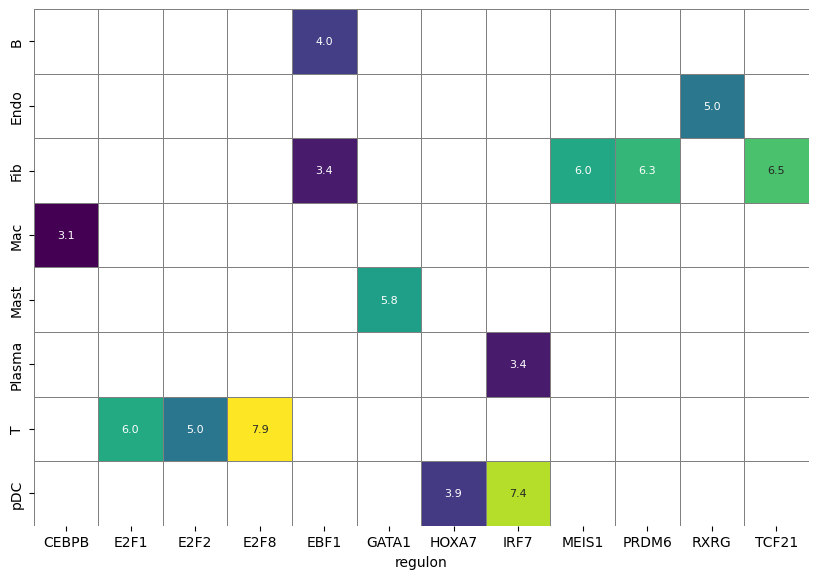
from pyscenic.rss import regulon_specificity_scores