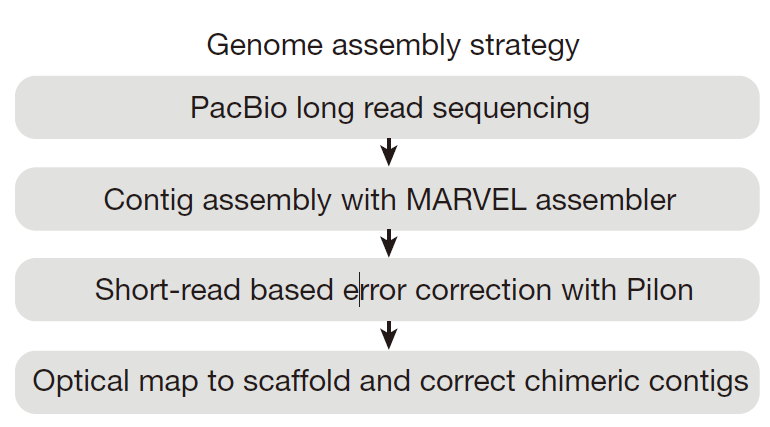
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
|
##!/usr/bin/env python
import multiprocessing
import marvel
import marvel.config
import marvel.queue
###### settings
DB = "ECOL" ## database name,需修改,和DBprepare.py中命名一样
COVERAGE = 25 ## coverage of the dataset,需修改
DB_FIX = DB + "_FIX" ## name of the database containing the patched reads
PARALLEL = multiprocessing.cpu_count() ## number of available processors
###### patch raw reads
q = marvel.queue.queue(DB, COVERAGE, PARALLEL)
###### run daligner to create initial overlaps
q.plan("{db}.dalign.plan")
###### run LAmerge to merge overlap blocks
q.plan("{db}.merge.plan")
## create quality and trim annotation (tracks) for each overlap block
q.block("{path}/LAq -b {block} {db} {db}.{block}.las")
## merge quality and trim tracks
q.single("{path}/TKmerge -d {db} q")
q.single("{path}/TKmerge -d {db} trim")
## run LAfix to patch reads based on overlaps
q.block("{path}/LAfix -g -1 {db} {db}.{block}.las {db}.{block}.fixed.fasta")
## join all fixed fasta files
q.single("!cat {db}.*.fixed.fasta > {db}.fixed.fasta")
## create a new Database of fixed reads (-j numOfThreads, -g genome size)
q.single("{path_scripts}/DBprepare.py -s 50 -r 2 -j 4 -g 4600000 {db_fixed} {db}.fixed.fasta", db_fixed = DB_FIX)
## run the commands
q.process()
########## assemble patched reads
q = marvel.queue.queue(DB_FIX, COVERAGE, PARALLEL)
###### run daligner to create overlaps
q.plan("{db}.dalign.plan")
###### run LAmerge to merge overlap blocks
q.plan("{db}.merge.plan")
###### start scrubbing pipeline
########## for larger genomes (> 100MB) LAstitch can be run with the -L option (preload reads)
########## with the -L option two passes over the overlap files are performed:
########## first to buffer all reads and a second time to stitch them
########## otherwise the random file access can make LAstitch pretty slow.
########## Another option would be, to keep the whole db in cache (/dev/shm/)
q.block("{path}/LAstitch -f 50 {db} {db}.{block}.las {db}.{block}.stitch.las")
########## create quality and trim annotation (tracks) for each overlap block and merge them
q.block("{path}/LAq -s 5 -T trim0 -b {block} {db} {db}.{block}.stitch.las")
q.single("{path}/TKmerge -d {db} q")
q.single("{path}/TKmerge -d {db} trim0")
########## create a repeat annotation (tracks) for each overlap block and merge them
q.block("{path}/LArepeat -c {coverage} -b {block} {db} {db}.{block}.stitch.las")
q.single("{path}/TKmerge -d {db} repeats")
########## detects "borders" in overlaps due to bad regions within reads that were not detected
########## in LAfix. Those regions can be quite big (several Kb). If gaps within a read are
########## detected LAgap chooses the longer part oft the read as valid range. The other part(s) are
########## discarded
########## option -L (see LAstitch) is also available
q.block("{path}/LAgap -t trim0 {db} {db}.{block}.stitch.las {db}.{block}.gap.las")
########## create a new trim1 track, (trim0 is kept)
q.block("{path}/LAq -s 5 -u -t trim0 -T trim1 -b {block} {db} {db}.{block}.gap.las")
q.single("{path}/TKmerge -d {db} trim1")
########## based on different filter critera filter out: local-alignments, repeat induced-alifnments
########## previously discarded alignments, ....
########## -r repeats, -t trim1 ... use repeats and trim1 track
########## -n 500 ... overlaps must be anchored by at least 500 bases (non-repeats)
########## -u 0 ... overlaps with unaligned bases according to the trim1 interval are discarded
########## -o 2000 ... overlaps shorter than 2k bases are discarded
########## -p ... purge overlaps, overlaps are not written into the output file
########## option -L (see LAstitch) is also available
q.block("{path}/LAfilter -n 300 -r repeats -t trim1 -T -o 2000 -u 0 {db} {db}.{block}.gap.las {db}.{block}.filtered.las")
########## merge all filtered overlap files into one overlap file
q.single("{path}/LAmerge -S filtered {db} {db}.filtered.las")
########## create overlap graph
q.single("{path}/OGbuild -t trim1 {db} {db}.filtered.las {db}.graphml")
########## tour the overlap graph and create contigs paths
q.single("{path_scripts}/OGtour.py -c {db} {db}.graphml")
q.single("{path}/LAcorrect -j 4 -r {db}.tour.rids {db} {db}.filtered.las {db}.corrected")
q.single("{path}/FA2db -c {db}_CORRECTED [expand:{db}.corrected.*.fasta]")
########## create contig fasta files
q.single("{path_scripts}/tour2fasta.py -c {db}_CORRECTED -t trim1 {db} {db}.tour.graphml {db}.tour.paths")
###### optional: create a layout of the overlap graph which can viewed in a Browser (svg) or Gephi (dot)
## q.single("{path}/OGlayout -R {db}.tour.graphml {db}.tour.layout.svg")
q.single("{path}/OGlayout -R {db}.tour.graphml {db}.tour.layout.dot")
q.process()
|